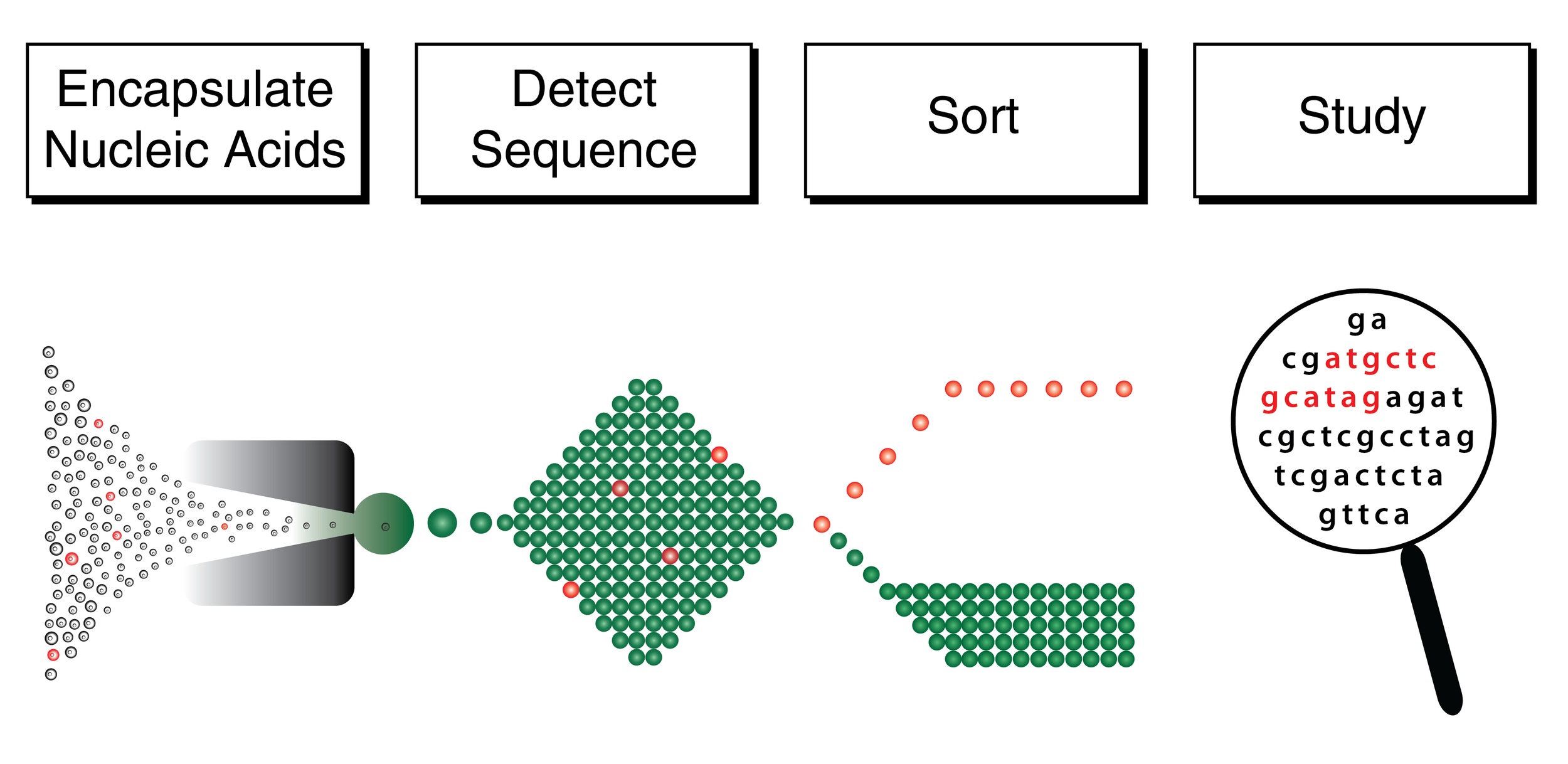
Nucleic Acid Cytometry
Nucleic Acid Cytometry (NAC) is a microdroplet technology for the enrichment of target DNA or RNA sequences. Nucleic acid fragments are encapsulated into millions of microdroplets; those carrying the target "keyword" (e.g. sequence) will be identified and sorted for downstream sequencing. NAC requires low amounts of input DNA and enables the enrichment of hundreds of kilobases flanking the target region. This is especially suitable for the study of genetic variation as well as targeted comparative metagenomics.
The general steps in a nucleic acid cytometry workflow (adapted from Clark et al. 2017).
Selected Publications
"Identification and genetic analysis of cancer cells with PCR-activated cell sorting"
D. J. Eastburn, A. Sciambi, and A. R. Abate
Nucleic Acids Research (2014)
"Microfluidic droplet enrichment for targeted sequencing"
D. J. Eastburn, Y. Huang, M. Pellegrino, A. Sciambi, L. J. Ptáček and A. R. Abate
Nucleic Acids Research (2015)
"Finding a helix in a haystack: nucleic acid cytometry with droplet microfluidics"
I. C. Clark and A. R. Abate
Lab on a Chip, DOI: 10.1039/C7LC00241F, Critical Review
"Sequence specific sorting of DNA molecules with FACS using 3dPCR"
D. J. Sukovich, S. T. Lance, and A. R. Abate
Scientific Reports, DOI: 10.1038/srep39385