Single-Cell Analysis
DAb-seq
DAb-seq is a multiomic platform combining targeted genotyping and immunophenotyping at the single-cell level. Through the use of DNA-antibody conjugates, phenotypic signal is encoded into next-generation sequencing data, providing a readout analogous to that of flow cytometry. The result is a dataset of linked proteogenomic information from thousands of single cells.
The experimental DAb-seq workflow, from sample collection to bioinformatics.
SiC-seq
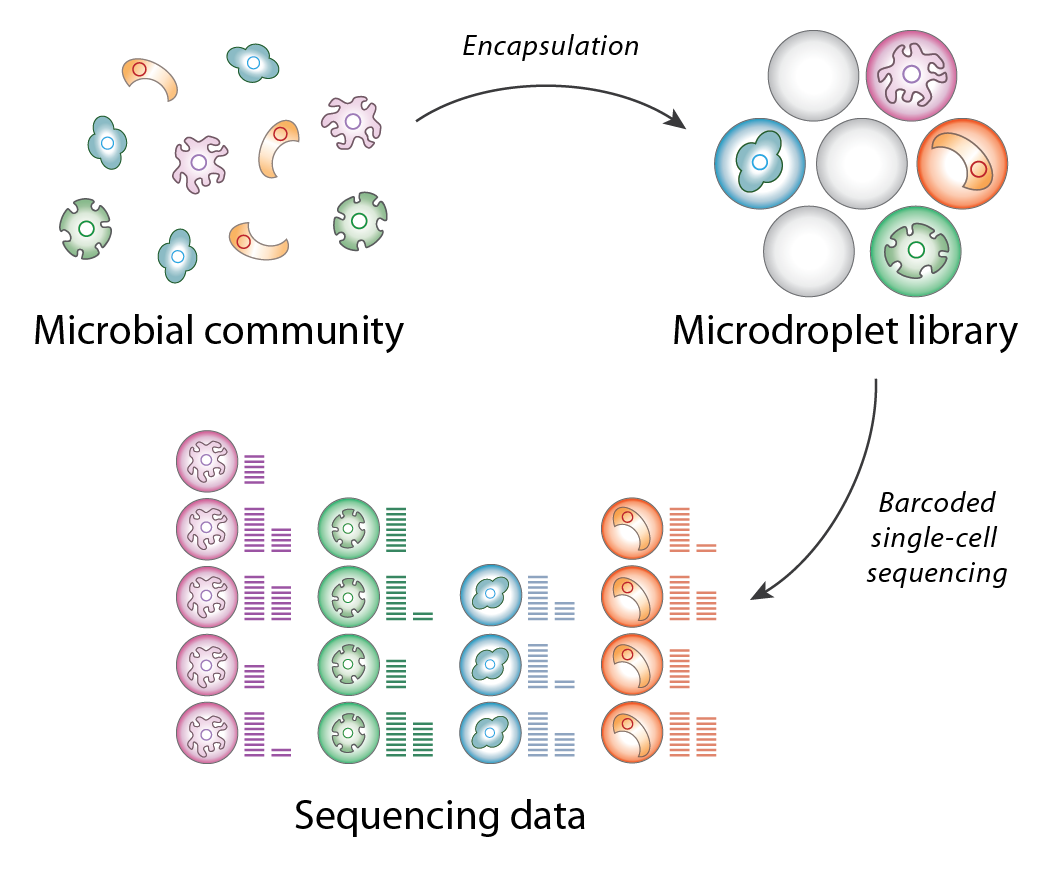
SiC-seq (Single-Cell sequencing) is a single-cell microfluidic platform for sequencing the genomes of tens of thousands of cells simultaneously at low coverage. In this workflow, cells from a metagenomic sample are encapsulated in micron-scale hydrogels and individually lysed, tagmented, and merged with a microdroplet containing a unique nucleic acid barcode, which is spliced onto the cell’s genomic DNA via single overlap extension PCR. This innovation in single-cell sequencing technology enables low-coverage genome sequencing of tens of thousands of bacterial cells in a single experiment.
Overview of the SiC-seq workflow
The primary applications of SiC-seq technology are in metagenomics, the study of the structure and function of microbial communities. Whereas metagenomic studies typically employ bulk methods such as shotgun or 16S sequencing to sample a community, SiC-seq compartmentalizes and barcodes each cell individually, preserving information from rare genotypes and closely-related strains.
A primary research direction is applying SiC-seq to de novo assembly of novel genomes from environmental samples. By clustering barcoded cells according to their genetic signatures, we can construct more accurate and complete genome assemblies than with shotgun sequencing alone.
Another aim for this project is to explore the mechanisms of genetic exchange between cells within a microbial community. Each SiC-seq experiment generates a database of sequencing information linked to many single cells, which can be mined for phylogenetic markers, functional genes, or any genomic features of interest. We call this method in silico cytometry. For example, this approach can be used to mine for phage sequence within thousands of cells simultaneously to develop maps of the phageome and infer patterns of viral transduction within a microbial community.
Abseq
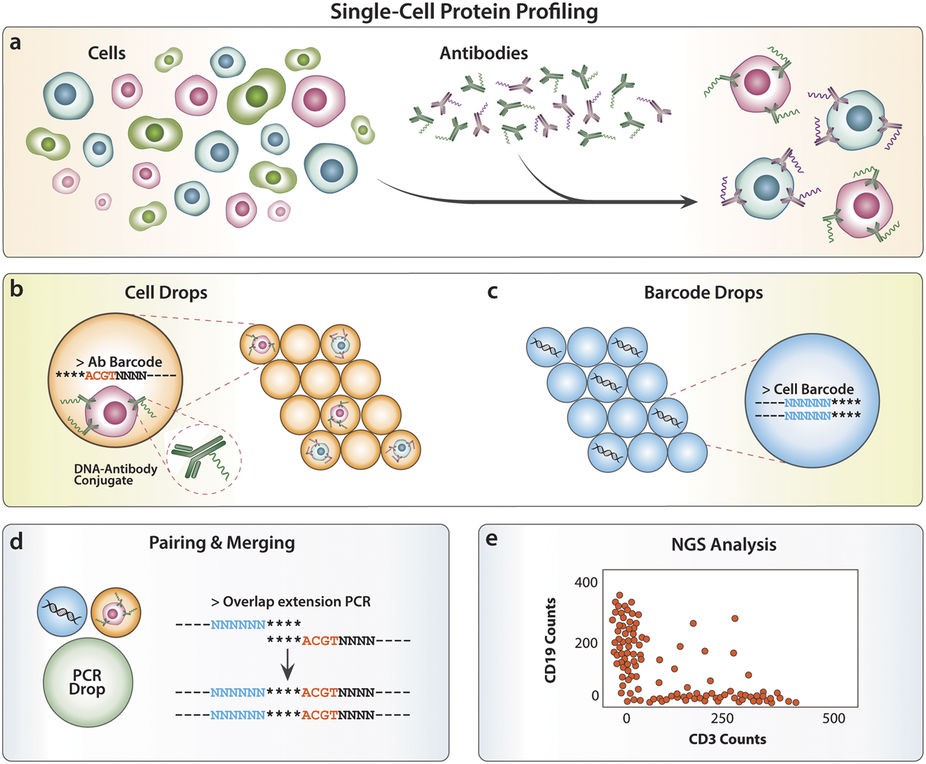
Abseq is a method to detect and quantitate proteins in single cells at ultrahigh throughput. Like flow and mass cytometry, Abseq uses specific antibodies to detect epitopes of interest; however, unlike these methods, antibodies are labeled with DNA sequence tags that can be read out with microfluidic barcoding and next-gen sequencing technologies. We demonstrate this novel approach by characterizing surface proteins of different cell types at the single-cell level and distinguishing between the cells by their protein expression profiles.
The Abseq experimental workflow for profiling single cells based on distinct cell surface proteins (from Shahi et al. 2017)
Selected Publications
"Single-cell genome sequencing at ultra-high-throughput with microfluidic droplet barcoding"
F. Lan, B. Demaree, N. Ahmed, and A.R. Abate
Nature Biotechnology, DOI: 10.1038/nbt.3880
"Abseq: Ultrahigh-throughput single cell protein profiling with droplet microfluidic barcoding"
P. Shahi, S. C. Kim, J. R. Haliburton, Z. J. Gartner & A. R. Abate
Scientific Reports, DOI: 10.1038/srep44447